Molecular docking study of modified isoniazid compounds on mycolic acid synthase in the cell wall of mycobacterium tuberculosis
Authors
Jordi Buannata , Bambang Wijianto , Ihsanul AriefDOI:
10.29303/aca.v7i2.173Published:
2024-10-31Issue:
Vol. 7 No. 2 (2024)Keywords:
molecular docking, Antituberculosis, Modified isoniazid compounds, Autodock VINA, ProTox-IIArticles
Downloads
How to Cite
Downloads
Metrics
Abstract
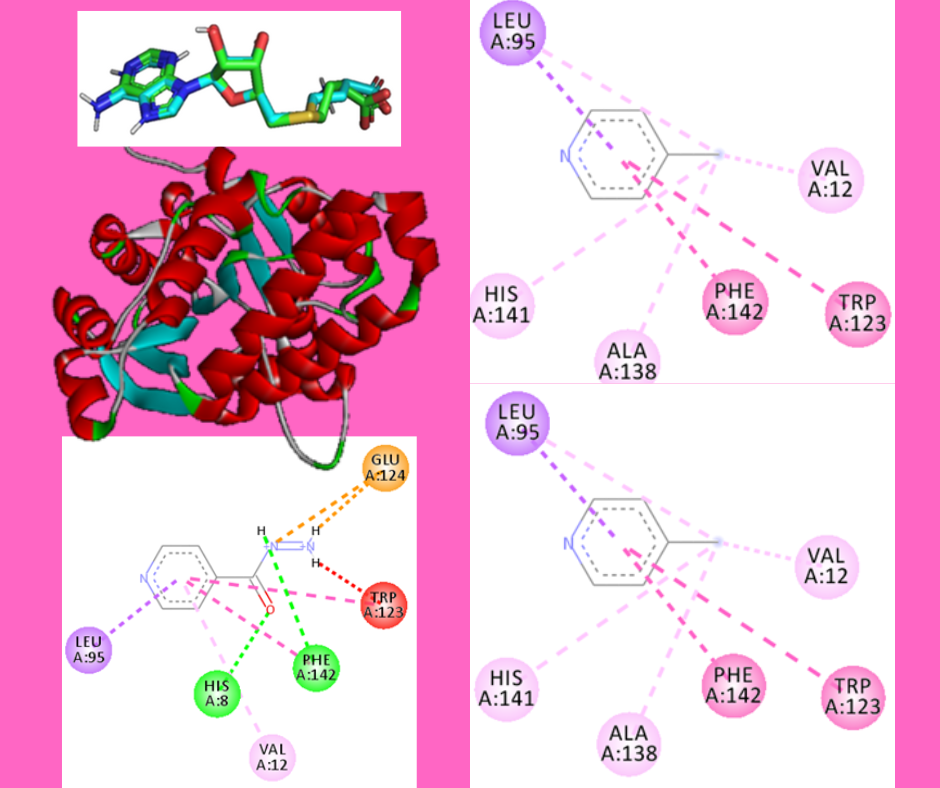
Using the isoniazid in antituberculosis therapy can lead to mutations in the KatG and inhA genes of Mycobacterium tuberculosis, resulting in the development of resistance and necessitating modifications to the isoniazid compound. This study aims to assess the potential and level of toxicity of modified compounds, namely 4-pyridine carboxylic acid, pyridine aldehyde, and methyl pyridine, on the mycolic acid receptor through a molecular docking approach. PyRx was employed for the docking process using a protocol with an exhaustiveness of 106 and a center grid box at X=42.424, Y=22.4321, and Z=46.6391. Additionally, the ProTox-II website was used to determine the toxicity level of the test compounds. The results obtained from this research consist of the respective affinity values of the test compounds: -6, -5.4, and -5.2 kcal/mol. The toxicity levels of the test compounds are as follows: class 5, class 4, and class 4. All test compounds interact with amino acids on the target protein, specifically with residue numbers Histidine (HIS A:8), Phenylalanine (PHE A:142) through hydrogen bonding, Leucine (LEU A:95) through pi-Sigma (π) bonding, and Valine (VAL A:12) through pi-Alkyl (π) bonding. In conclusion, the 4-pyridine carboxylic acid compound exhibits potential as a promising drug candidate but comes with a high level of toxicity
References
Venkatappa, T., Shen, D., Ayala, A., Li, R., Sorri, Y., Punnoose, R., Katz, D. (2023).: Association of Mycobacterium tuberculosis infection test results with risk factors for tuberculosis transmission. Journal of Clinical Tuberculosis and Other Mycobacterial Diseases. 33, 100386. https://doi.org/10.1016/j.jctube.2023.100386.
Sampiron, E.G., Calsavara, L.L., Baldin, V.P., Montaholi, D.C., Leme, A.L.D., Namba, D.Y., Alves Olher, V.G., Caleffi-Ferraciolli, K.R., Cardoso, R.F., Siqueira, V.L.D., Vandresen, F., Scodro, R.B. de L. (2023). Isoniazid-N-acylhydrazones as promising compounds for the anti-tuberculosis treatment. Tuberculosis. 141, 102363. https://doi.org/10.1016/j.tube.2023.102363
Chan, C.-Y., Au-Yeang, C., Yew, W.-W., Hui, M., Cheng, A.F.B. (2001). Postantibiotic Effects of Antituberculosis Agents Alone and in Combination. Antimicrob Agents Chemother. 45, 3631–3634. https://doi.org/10.1128/AAC.45.12.3631-3634.2001
Wang Kun, Deng Yimin, Cui Xujie, Chen Mengli, Ou Yanzhe, Li Danting, Guo Minhao, Li Weihui. (2023). PatA Regulates Isoniazid Resistance by Mediating Mycolic Acid Synthesis and Controls Biofilm Formation by Affecting Lipid Synthesis in Mycobacteria. Microbiology Spectrum. 11, e00928-23. https://doi.org/10.1128/spectrum.00928-23
North, E., Jackson, M., Lee, R. (2013). New Approaches to Target the Mycolic Acid Biosynthesis Pathway for the Development of Tuberculosis Therapeutics. CPD. 20, 4357–4378. https://doi.org/10.2174/1381612819666131118203641
Holzheimer, M., Buter, J., Minnaard, A.J. (2021). Chemical Synthesis of Cell Wall Constituents of Mycobacterium tuberculosis. Chem. Rev. 121, 9554–9643. https://doi.org/10.1021/acs.chemrev.1c00043.
PaweŁczyk Jakub, Kremer Laurent. (2014). The Molecular Genetics of Mycolic Acid Biosynthesis. Microbiology Spectrum. 2, 10.1128/microbiolspec.mgm2-0003–2013. https://doi.org/10.1128/microbiolspec.mgm2-0003-2013
Takayama, K., Wang, C., Besra, G.S. (2005). Pathway to Synthesis and Processing of Mycolic Acids in Mycobacterium tuberculosis. Clin Microbiol Rev. 18, 81–101. https://doi.org/10.1128/CMR.18.1.81-101.2005.
Dyas, R.A.A., Wijianto, B., Ih, H. (2023). Docking studies for screening antibacterial compounds of Red Jeringau (Acorus calamus L.) using Shigella flexneri protein as a model system. Acta.Chim.Asiana. 6, 343–350. https://doi.org/10.29303/aca.v6i2.161
Budiarto, D., Wijianto, B., Ih, H. (2023). Study of Anthocyanin Molecule Blocking as Anti-Hypertensive through the Pathway of the Renin-Angiotensin-Aldosterone System (RAAS). Indo. J. Chem. Res. 11, 49–58. https://doi.org/10.30598//ijcr.2023.11-bud
Wijianto, B., Ritmaleni, R., Hari, P., Arief, N. (2020). In silico and in vitro anti-inflammatory evaluation of 2,6-bis-(3’-ethoxy, 4’-hydroxybenzylidene)-cyclohexanone, 2,6-bis-(3’-Bromo,4’-methoxybenzylidene)-cyclohexanone, and 2,6-bis- (3’,4’-dimethoxybenzylidene)-cyclohexanone. J app pharm sci. 10, 99–106. https://doi.org/10.7324/JAPS.2020.10613.
Wijianto, B., . R., Purnomo, H., Nurrochmad, A. (2019). In silico and in vitro assay of HGV analogue as antibacterial. Int J Pharm Pharm Sci. 78–85. https://doi.org/10.22159/ijpps.2019v11i3.30581.
C Malau, N. D., & Azzahra, S. F. (2020). Analysis docking of plasmodium falciparum enoyl acyl carrier protein reductase (pfenr) with organic compunds from virtual screening of herbal database. Acta Chimica Asiana, 3(1), 127-134. Dyas, R. A. A., Wijianto, B., & Hariyanto, I. H. (2023). Docking studies for screening antibacterial compounds of Red Jeringau (Acorus calamus L.) using Shigella flexneri protein as a model system. Acta Chimica Asiana, 6(2), 343-350. https://doi.org/10.4081/jphia.2023.2532
Banerjee, P., Eckert, A.O., Schrey, A.K., Preissner, R. (2018). ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Research. 46, W257–W263. https://doi.org/10.1093/nar/gky318
Cavasotto, C.N., Scardino, V. (2022). Machine Learning Toxicity Prediction: Latest Advances by Toxicity End Point. ACS Omega. 7, 47536–47546. https://doi.org/10.1021/acsomega.2c05693
Yang, S., Kar, S. (2023). Application of artificial intelligence and machine learning in early detection of adverse drug reactions (ADRs) and drug-induced toxicity. Artificial Intelligence Chemistry. 1, 100011. https://doi.org/10.1016/j.aichem.2023.100011
Pires, D.E.V., Blundell, T.L., Ascher, D.B. (2015). pkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. J. Med. Chem. 58, 4066–4072. https://doi.org/10.1021/acs.jmedchem.5b00104
Wade, A.D., Huggins, D.J. (2020). Identification of Optimal Ligand Growth Vectors Using an Alchemical Free-Energy Method. J. Chem. Inf. Model. 60, 5580–5594. https://doi.org/10.1021/acs.jcim.0c00610
Du, X., Li, Y., Xia, Y.-L., Ai, S.-M., Liang, J., Sang, P., Ji, X.-L., Liu, S.-Q. (2016). Insights into Protein–Ligand Interactions: Mechanisms, Models, and Methods. IJMS. 17, 144. https://doi.org/10.3390/ijms17020144
Decherchi, S., Cavalli, A. (2020). Thermodynamics and Kinetics of Drug-Target Binding by Molecular Simulation. Chem. Rev. 120, 12788–12833. https://doi.org/10.1021/acs.chemrev.0c00534
Kim, S., Thiessen, P.A., Bolton, E.E., Chen, J., Fu, G., Gindulyte, A., Han, L., He, J., He, S., Shoemaker, B.A., Wang, J., Yu, B., Zhang, J., Bryant, S.H. (2016). PubChem Substance and Compound databases. Nucleic Acids Res. 44, D1202–D1213. https://doi.org/10.1093/nar/gkv951
Kim, S., Chen, J., Cheng, T., Gindulyte, A., He, J., He, S., Li, Q., Shoemaker, B.A., Thiessen, P.A., Yu, B., Zaslavsky, L., Zhang, J., Bolton, E.E. (2023). PubChem 2023 update. Nucleic Acids Research. 51, D1373–D1380. https://doi.org/10.1093/nar/gkac956
Tanbin, S., Ahmad Fuad, F.A., Abdul Hamid, A.A. (2020). Virtual Screening for Potential Inhibitors of Human Hexokinase II for the Development of Anti-Dengue Therapeutics. BioTech. 10, 1. https://doi.org/10.3390/biotech10010001
License
Copyright (c) 2024 Jordi Buannata, Bambang Wijianto, Ihsanul Arief

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
Authors who publish with ACA: Acta Chimica Asiana agree to the following terms:
- Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. This license allows authors to use all articles, data sets, graphics, and appendices in data mining applications, search engines, web sites, blogs, and other platforms by providing an appropriate reference. The journal allows the author(s) to hold the copyright without restrictions and will retain publishing rights without restrictions.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgement of its initial publication in ACA: Acta Chimica Asiana.
- Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See The Effect of Open Access).





 Indonesian Chemical Society, Chapter Nusa Tenggara. Jalan Majapahit 62 Mataram, University of Mataram, 83125, Indonesia
Indonesian Chemical Society, Chapter Nusa Tenggara. Jalan Majapahit 62 Mataram, University of Mataram, 83125, Indonesia





