Docking studies for screening antibacterial compounds of Red Jeringau (Acorus calamus L.) using Shigella flexneri protein as a model system
Authors
Riyadh Aqilsya Amaryl Dyas , Bambang Wijianto , Hariyanto IHDOI:
10.29303/aca.v6i2.161Published:
2023-06-24Issue:
Vol. 6 No. 2 (2023)Keywords:
Red Jeringau (Acorus calamus L.), α and β-Asarone, Antibacterial, Autodock VINA, ProTox-II5Articles
Downloads
How to Cite
Downloads
Metrics
Abstract
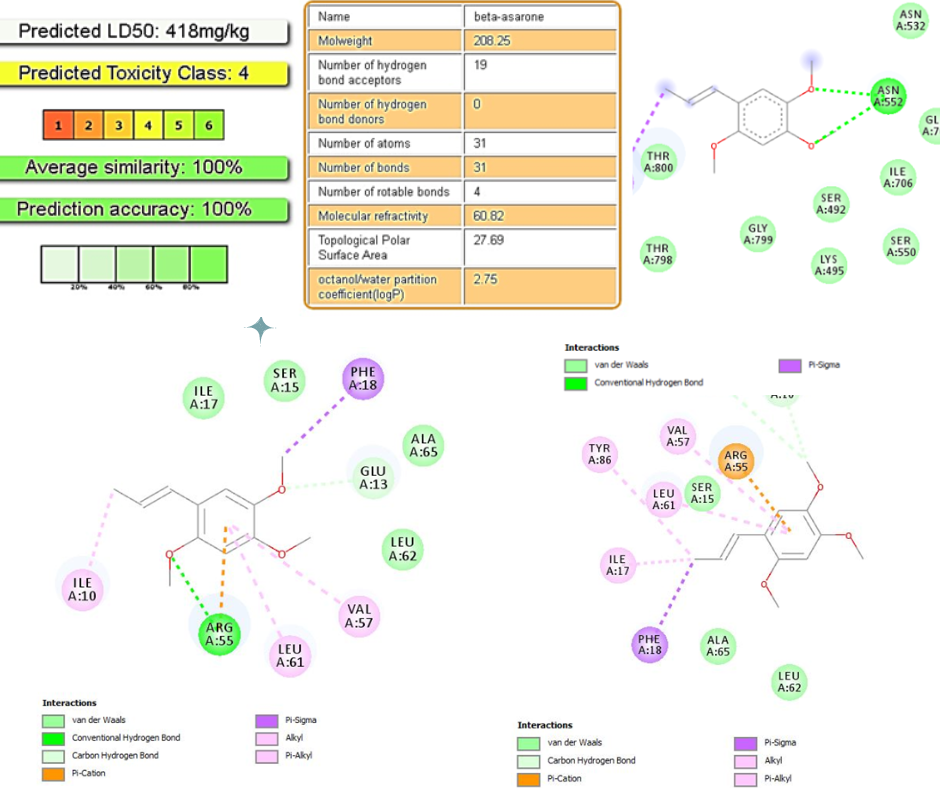
Alpha (a) and beta (β) asarone were identified as the main compounds of red Jeringau (Acorus calamus L.) and had antimicrobial properties. This study aimed to know these two compounds' antibacterial mechanism and toxicity prediction against the PBP 2 protein and 50S Ribosomal Protein of Shigella flexneri. Molecular docking protocol using PyRx device was performed with Exhaustiveness value= 106, grid x=38.738375, y=112.645792, z=46.926417 for PBP2, and grid x=71.721251, y=47.551601, z=9.663173 for 50S Ribosomal Protein. The molecular docking results on the α -Asarone compound obtained an affinity value of -5.7 kcal/mol for PBP2 and an affinity value of -5.6 kcal/mol for 50S Ribosomal Protein. In comparison, β-Asarone had an affinity value of -5.6 kcal/mol to PBP2 and an affinity value of -5.7 kcal/mol for 50S Ribosomal Protein. The α and β-Asarone affinity had better values than the control. Molecular docking of α and β-Asarone compounds results in ionic bonds to the TYR529 amino acid and polar bonds to the ASN552 amino acid of PBP2. However, only β-Asarone produces ionic bonds at the amino acid ILE17 and polar bonds at GLU13 from 50S Ribosomal Protein. Based on this study, the α and β-Asarone compounds were shown to have antibacterial activity by interfering with the permeability of the bacterial cell wall. Both compounds are also predicted to have carcinogenic and mutagen effects.
References
Schnupf Pamela, Sansonetti Philippe J. (2019). Shigella Pathogenesis: New Insights through Advanced Methodologies. Microbiol. Spectr. 7, 7.2.28. doi:10.1128/microbiolspec.BAI-0023-2019.
Baker, S., The, H.C. (2018). Recent insights into Shigella. Curr Opin Infect Dis. 31, 449–454. doi:10.1097/QCO.0000000000000475.
Williams, P.C.M., Berkley, J.A. (2018). Guidelines for the treatment of dysentery (shigellosis): a systematic review of the evidence. Paediatr Int Child Health. 38, S50–S65. doi:10.1080/20469047.2017.1409454.
Pakbin, B., Brück, W.M., Brück, T.B. (2023). Molecular Mechanisms of Shigella Pathogenesis; Recent Advances. Int. J. Mol. Sci. 24. doi: 10.3390/ijms24032448.
Miti, S., Chilyabanyama, O.N., Chisenga, C.C., Chibuye, M., Bosomprah, S., Mumba, C., Chitondo, S., Siziya, S., Cohen, D., Chilengi, R., Simuyandi, M. (2023). Sensitivity and predictive value of dysentery in diagnosing shigellosis among under five children in Zambia. PLoS One. 18, e0279012–e0279012. doi:10.1371/journal.pone.0279012.
Nasser, A., Mosadegh, M., Azimi, T., Shariati, A. (2022). Molecular mechanisms of Shigella effector proteins: a common pathogen among diarrheic pediatric population. Mol Cell Pediatr. 9, 12–12. doi:10.1186/s40348-022-00145-z.
Gharpure, R., Marsh, Z.A., Tack, D.M., Collier, S.A., Strysko, J., Ray, L., Payne, D.C., Garcia-Williams, A.G. (2021). Disparities in Incidence and Severity of Shigella Infections Among Children-Foodborne Diseases Active Surveillance Network (FoodNet), 2009-2018. J Pediatric Infect Dis Soc. 10, 782–788. doi:10.1093/jpids/piab045.
Bowen, A., Eikmeier, D., Talley, P., Siston, A., Smith, S., Hurd, J., Smith, K., Leano, F., Bicknese, A., Norton, J.C., Campbell, D., Centers for Disease Control and Prevention (CDC). (2015). Notes from the Field: Outbreaks of Shigella sonnei Infection with Decreased Susceptibility to Azithromycin Among Men Who Have Sex with Men - Chicago and Metropolitan Minneapolis-St. Paul, 2014. MMWR Morb Mortal Wkly Rep. 64, 597–598.
Ranganathan, S., Doucet, M., Grassel, C.L., Delaine-Elias, B., Zachos, N.C., Barry, E.M. (2019). Evaluating Shigella flexneri Pathogenesis in the Human Enteroid Model. Infect Immun. 87, e00740-18. doi:10.1128/IAI.00740-18.
Kelmani R, C. (2018). Shigellosis: Its Prevention and Management Issues. CDVS. 1. doi:10.32474/CDVS.2018.01.000121.
Ranjbar, R., Farahani, A. (2019). Shigella: Antibiotic-Resistance Mechanisms and New Horizons for Treatment. Infect Drug Resist. 12, 3137–3167. doi:10.2147/IDR.S219755
Muchtaromah, B., Hayati, A., Agustina, E. (2019). Phytochemical Screening and Antibacterial Activity of Acorus calamus L. Extracts. Jurnal Biodjati. 4, 68–78. doi:10.15575/biodjati.v4i1.4235.
Rita, W.S., Swantara, I.M.D., Utami, G.A.P. (2019). Antimicrobial activity of Acorus calamus L. rhizome extract and its total flavonoid and phenolic contents. AIP Conference Proceedings. 2155, 020054. doi:10.1063/1.5125558.
Wijianto, B., Ritmaleni, R., Purnomo, H., Nurrochmad, A. (2019). In silico and in vitro assay of HGV analogue as antibacterial. Int J Pharm Pharm Sci. 11, 78–85. doi:10.22159/ijpps.2019v11i3.30581.
Wijianto, B., Ritmaleni, R., Purnomo, H., Nurrochmad, A. (2020). In silico and in vitro anti-inflammatory evaluation of 2,6-bis-(3’-ethoxy, 4’-hydroxybenzylidene)-cyclohexanone, 2,6-bis-(3’-Bromo,4’-methoxybenzylidene)-cyclohexanone, and 2,6-bis- (3’,4’-dimethoxybenzylidene)-cyclohexanone. J app pharm sci. 10, 99–106. doi:10.7324/JAPS.2020.10613.
Wijianto, B., Ritmaleni, Purnomo, H., Nurrochmad, A. (2020). Curcumin mono-carbonyl analogs as potent antibacterial compounds: synthesis, biological evaluation and docking simulation study. RJC. 13, 1153–1165. doi:10.31788/RJC.2020.1325554.
Eberhardt, J., Santos-Martins, D., Tillack, A.F., Forli, S. (2021). AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 61, 3891–3898. doi: 10.1021/acs.jcim.1c00203.
Zapun, A., Contreras-Martel, C., Vernet, T. (2008). Penicillin-binding proteins and β-lactam resistance. FEMS Microbiology Reviews. 32, 361–385. doi: 10.1111/j.1574-6976.2007.00095.x.
Watkins, R.R., Bonomo, R.A. (2017). 140 - β-Lactam Antibiotics. In: Cohen, J., Powderly, W.G., and Opal, S.M. (eds.) Infectious Diseases (Fourth Edition). pp. 1203-1216.e2. Elsevier.
López-Jacome, L.E., Mercado-Casillas, Y.M., Méndez-Sotelo, B.J., Jiménez-Cortes, J.G., Tovar-García, A., Estrada-Velasco, A.Y., Almeida-Villegas, J.A., Martínez, J.D.P., García-Contreras, R. (2022). Anti-Bacterial Agents. In: Rezaei, N. (ed.) Encyclopedia of Infection and Immunity. pp. 494–509. Elsevier, Oxford.
Trott, O., Olson, A.J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 31, 455–461. doi:10.1002/jcc.21334.
Wang, R., Lai, L., Wang, S. (2002). Further development and validation of empirical scoring functions for structure-based binding affinity prediction. J. Comput. Aided Mol. 16, 11–26. doi:10.1023/A:1016357811882.
Hassan, N.M., Alhossary, A.A., Mu, Y., Kwoh, C.-K.: Protein-Ligand Blind Docking Using QuickVina-W With Inter-Process Spatio-Temporal Integration. Scientific Reports. 7, 15451 (2017). doi:10.1038/s41598-017-15571-7.
Quiroga, R., Villarreal, M.A. (2016). Vinardo: A Scoring Function Based on Autodock Vina Improves Scoring, Docking, and Virtual Screening. PLOS ONE. 11, e0155183. doi:10.1371/journal.pone.0155183.
Tang, S., Chen, R., Lin, M., Lin, Q., Zhu, Y., Ding, J., Hu, H., Ling, M., Wu, J. (2022). Accelerating AutoDock Vina with GPUs. Molecules. 27. doi:10.3390/molecules27093041.
Wijianto, B., Hamzah, H., Nurhidayah A. L., Kemuning G. I., Dyas, R. A. A,. (2022). Characterization of Onchidiid Slug (Onchidium typhae) West Kalimantan Waters as Antibacterials and Antifungal. Borneo Journal of Pharmacy. 5, 35–41. doi:10.33084/bjop.v5i1.2936.
Zoete, V., Grosdidier, A., Michielin, O. (2009). Docking, virtual high throughput screening and in silico fragment-based drug design. J Cell Mol Med. 13, 238–248. doi:10.1111/j.1582-4934.2008.00665.x.
Morris, G.M., Huey, R., Lindstrom, W., Sanner, M.F., Belew, R.K., Goodsell, D.S., Olson, A.J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791. doi:10.1002/jcc.21256.
Hill, A.D., Reilly, P.J. (2015). Scoring Functions for AutoDock. In: Lütteke, T. and Frank, M. (eds.) Glycoinformatics. pp. 467–474. Springer New York, New York, NY
Wu, D., Karimi-Maleh, H., Liu, X., Fu, L.: Bibliometrics Analysis of Research Progress of Electrochemical Detection of Tetracycline Antibiotics. J. Anal. Chem. 2023, 6443610 (2023). https://doi.org/10.1155/2023/6443610
Banerjee, P., Eckert, A.O., Schrey, A.K., Preissner, R. (2018). ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 46, W257–W263. doi:10.1093/nar/gky318.
Cartus, A.T., Schrenk, D. (2016). Metabolism of the carcinogen alpha-asarone in liver microsomes. FCT. 87, 103–112. doi:10.1016/j.fct.2015.11.021.
Cartus, A.T., Stegmüller, S., Simson, N., Wahl, A., Neef, S., Kelm, H., Schrenk, D. (2015). Hepatic Metabolism of Carcinogenic β-Asarone. Chem. Res. Toxicol. 28, 1760–1773. doi:10.1021/acs.chemrestox.5b00223.
License
Copyright (c) 2023 Riyadh Aqilsya Amaryl Dyas, Bambang Wijianto, Hariyanto IH

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
Authors who publish with ACA: Acta Chimica Asiana agree to the following terms:
- Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. This license allows authors to use all articles, data sets, graphics, and appendices in data mining applications, search engines, web sites, blogs, and other platforms by providing an appropriate reference. The journal allows the author(s) to hold the copyright without restrictions and will retain publishing rights without restrictions.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgement of its initial publication in ACA: Acta Chimica Asiana.
- Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See The Effect of Open Access).





 Indonesian Chemical Society, Chapter Nusa Tenggara. Jalan Majapahit 62 Mataram, University of Mataram, 83125, Indonesia
Indonesian Chemical Society, Chapter Nusa Tenggara. Jalan Majapahit 62 Mataram, University of Mataram, 83125, Indonesia





